How Much Do You Know About the Prokaryotic Expression System?
2018-02-21
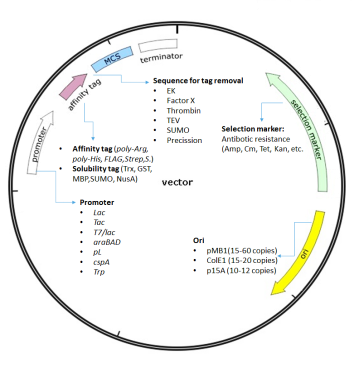
E. coli protein expression system is the most commonly used expression system for recombinant proteins. Prokaryotic expression vectors generally consist of the replicon, promoters, selection markers, multiple cloning sites (MCS) and fusion tags / restriction sites.
Replicon
The replicon is comprised of the origin of replication (ORI) and all of its control elements. The ORI is the place where DNA replication begins, enabling a plasmid to reproduce itself as it must to survive within cells. The best choice of ORI depends on how many plasmid copies you want to maintain, which host or hosts you intend to use, and whether or not you need to consider your plasmid's compatibility with one or more other plasmids. Commonly used pET (Novagen) series vectors mainly carry pMB1 ori, providing a copy number of 15-60; pQE vectors (Qiagen) carrying ColE1 ori provides a copy number of 15-20; pACYC and pBAD series of vectors carry p15A ori, giving a copy number of 10-12, and are commonly used for the co-expression of two recombinant proteins. (Misunderstanding: Does high copy number of the carrier result in high protein expression yield? High copy of the exogenous plasmids will create a greater burden on the normal metabolism of the host, which may slow down cell proliferation, destabilize the plasmid, and eventually compromise the final expression yield of the recombinant protein.) This following table defines common cloning vectors, their copy number, ORI, and incompatibility groups.
Table 1. Common Prokaryotic Expression Vectors and ORIs
|
Vectors
|
Copy Number |
ORI |
Incompatibility Group |
Control |
|---|---|---|---|---|
|
pUC |
~500-700 |
pMB1 (derivative) |
A |
Relaxed |
|
pBR322 |
~15-20 |
pMB1 |
A |
Relaxed |
|
pET |
~15-20 |
pBR322 |
A |
Relaxed |
|
pGEX |
~15-20 |
pBR322 |
A |
Relaxed |
|
pColE1 |
~15-20 |
ColE1 |
A |
Relaxed |
|
pR6K |
~15-20 |
R6K* |
C |
Stringent |
|
pACYC |
~10 |
p15A |
B |
Relaxed |
|
pSC101 |
~5 |
pSC101 |
C |
Stringent |
|
pBluescript |
~300-500 |
ColE1 (derivative) and F1** |
A |
Relaxed |
|
pGEM |
~300-500 |
pUC and F1** |
A |
Relaxed |
Note the A -C compatibility grouping is an arbitrary designation, and plasmids from the same incompatibility group should not be co-transformed.
* Requires pir gene for replication.
**F1 is a phage-derived ORI that allows for the replication and packaging of ssDNA into phage particles. Plasmids with phage-derived ORIs are referred to as phagemid
Promoter
Refers to the DNA sequence that is specific for RNA polymerase’s recognition and binding. Table 2 illustrates a few of the most common prokaryotic expression promoters.
Table 2. Common Prokaryotic Expression Promoters
| Promoters | Description | Features |
|---|---|---|
| Lac |
Constitutive in the absence of lac repressor (lacI or lacIq). Can be induced by IPTG or lactose.
|
Weak expression due to lacI repression thus is not commonly used in high copy-number vector. Generally high constitutive expression
|
| T7lac |
Requires T7 RNA polymerase and inducible by IPTG. Commonly found in pET vectors
|
Most frequently used procryptic promoter. Target protein yield as high as 50% of total protein expression
|
| Tac |
Hybrid promoter containing -35 region from trpB and -10 region from lac, inducible by IPTG
|
Very tight regulation. Generally better expression level than lac alone
|
| Trp |
Promoter from E. coli tryptophan operon
|
Repressible, gets turned off with high levels of cellular tryptophan.
|
| araBAD |
Promoter of the arabinose metabolic operon, Inducible by arabinose
|
Commonly found in pBAD vectors. Good for rapid regulation and low basal expression
|
| pL |
Promoter from bacteriophage lambda. Can be regulated by temperature
|
Induction temp. at 40-42℃
|
| CspA |
Induction temp. at 15℃
|
election Marker
Prokaryotic expression system often uses antibiotics for screening. Commonly used antibiotics are ampicillin, kanamycin, tetracycline, chloramphenicol, etc.
Fusion Tags
Protein fusion tags can be roughly divided into affinity tags and solubility tags. Affinity tags are used primarily for protein purification by affinity chromatography. Solubility tags are often incorporated to promote proper protein folding and enhance solubility. Table 3 and Table 4 list some of the common affinity tags and solubility-promoting tags.
Table 3. Common Affinity Tags
|
Tag
|
Length (aa)
|
Matric
|
Purification
|
|---|---|---|---|
|
Poly-arginine
|
5 (RRRRR)
|
Cation exchange resin
|
0-400 mM NaCl linear elution |
|
Hexa-histidine
|
6 (HHHHHH)
|
Metal ions (Ni2+, Co, Cu, Zn, Fe3+ |
0-300 mM imidazole linear elution |
|
FLAG
|
8 (DYKDDDDK)
|
Anti-FLAG mAb
|
2-5 mM EDTA
|
|
Strep II
|
8 (WSHPQFEK)
|
Strep-Tactin (modified streptavidin) |
2-25mM desulfated biotin |
|
S
|
15 (KETAAAKFERQHMDS) |
S-protein of RNase A
|
3M guanidinium isothiocyanate or 0.2 M potassium citrate, pH 2.0 |
|
Halo
|
~300
|
HaloLink? Resin
|
|
Table 4. Common Solubility Tags
|
Solubility Tags
|
Length (aa)
|
Matric
|
Solubility Enhancement
|
|---|---|---|---|
|
Trx
|
109
|
Oftenco-expressed with His tag
|
+++
|
|
GST
|
211
|
Glutathione Sepharose
|
+
|
|
MBP
|
396
|
Cross-linked amylose
|
+++
|
|
SUMO
|
~100
|
Often co-expressed with His tag
|
++++
|
|
Nus
|
495
|
Often co-expressed with His tag
|
++
|
Label Removal
Protein tags are usually removed by enzymatic cleavage after the corresponding affinity purification or dissolution promotion is completed, as the presence of the tag may interfere with the normal function of the target protein. The tag and the recombinant protein are then separated by an affinity column, which simultaneously removes the recombinant endoprotease that often carries an affinity tag as well. N-terminal tags are generally easy to remove while the complete removal of the C-tag is more difficult because most proteases cleave only at the C-terminus of the recognition sequence. Table 5 lists some of the commonly used proteases and their restriction sites.
Table 5. Common Protaminase
|
Protease
|
Restriction Site
|
|---|---|
|
Enterokinase (EK enzyme)
|
DDDDK
|
|
Factor X
|
IEGRK
|
|
Thrombin
|
LVPRK
|
|
Prescission
|
LEVLFQ
|
|
TEV
|
ENLYFQ
|
|
SUMO(small ubiquitin-related modifier)
|
Identify the tertiary structure of the sumo sequence and cleave at the c-terminus of the conserved gly residue |
References:
1.Rosano, Germán L., and Eduardo A. Ceccarelli. "Recombinant protein expression in Escherichia coli: advances and challenges." Frontiers in microbiology 5 (2014).
2.Guide to protein purification[M]. Academic Press, 2009.

