 +1 302-235-9792
+1 302-235-9792

- News
- AI Shines with the Nobel Prize! High-throughput Antibody Expression Platform Fuels AI-Driven Next-Generation Antibody Discovery
AI Shines with the Nobel Prize! High-throughput Antibody Expression Platform Fuels AI-Driven Next-Generation Antibody Discovery
The 2024 Nobel Prize in Chemistry was awarded half to David Baker for his contributions to computational protein design, and the other half jointly to Demis Hassabis and John M. Jumper for their contributions to protein structure prediction. Demis Hassabis and John M. Jumper successfully utilized artificial intelligence technology to predict the structures of nearly all known proteins, while David Baker mastered the building blocks of life and created entirely new proteins.

AlphaFold: Fast and Accurate Protein Structure Prediction
A group of laureates, Demis Hassabis and John M. Jumper, were involved in creating the AI protein structure analysis tool AlphaFold.The function of a protein is determined by its three-dimensional spatial structure. Therefore, to truly understand how proteins function, scientists must accurately determine their spatial structure.
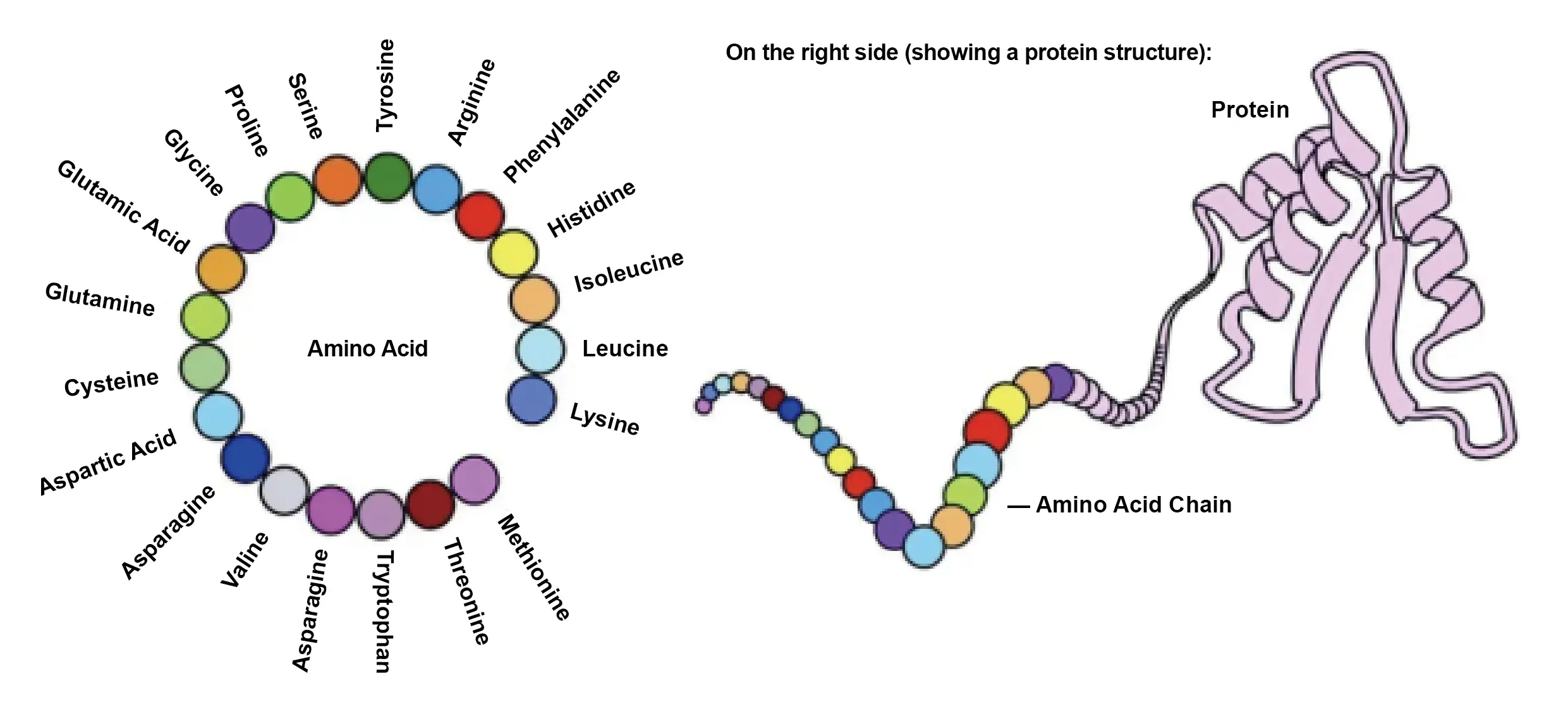
Proteins are typically composed of 20 different amino acids, but they can be combined in countless ways. The amino acid chain twists and folds into a unique (sometimes one-of-a-kind) three-dimensional structure, and it is this structure that gives the protein its function.
From the first generation in 2018 to the third generation in 2024, AlphaFold has undergone significant upgrades and improvements. Starting with the initial model that used a classic convolutional neural network architecture, to the second generation model that introduced the more powerful Transformer architecture and achieved unprecedented prediction accuracy, and then to the third generation model, which can predict the structures of protein complexes involving "almost all types of molecules," its capabilities have continuously advanced.
Determining Protein Structures Using AlphaFold2
Rosetta: Creating Completely New Proteins
Sharing the Nobel Prize with the two artificial intelligence researchers is biochemist David Baker, whose research focuses on using computer software to design entirely new proteins from scratch.
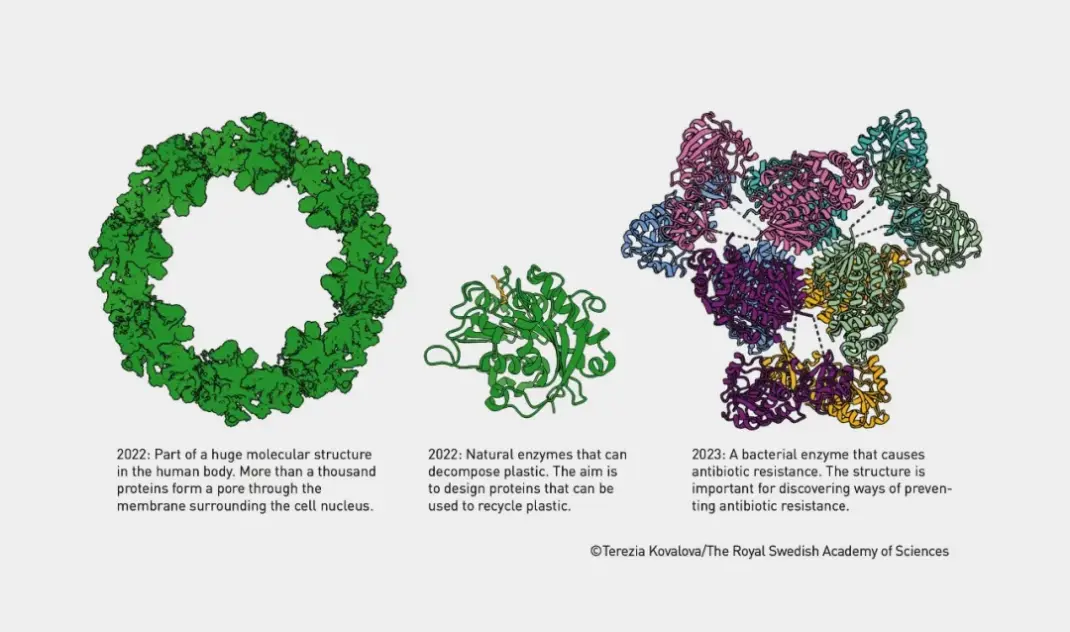
Professor David Baker has greatly advanced the field of protein structure prediction and design by developing the Rosetta software platform. Rosetta not only accurately predicts how amino acid chains fold into complex three-dimensional structures, but it also supports the design of new proteins that do not exist in nature. These new proteins can perform specific functions, such as catalyzing novel chemical reactions or serving as therapeutic drugs. Today, his research team continues to create imaginative proteins, many of whose structures can be used in fields like drugs, vaccines, nanomaterials, and micro-sensors.
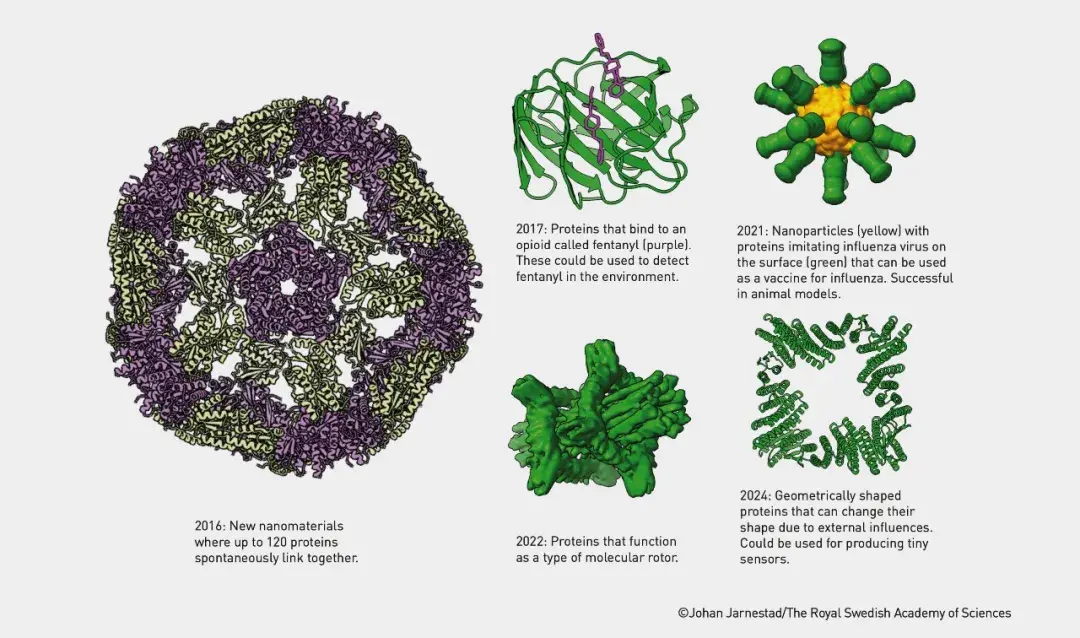
Proteins Built Using Rosetta
Currently, applying artificial intelligence methods to analyze data and construct powerful predictive and generative models for complex biological phenomena has become a major trend in technological innovation across various fields.
AI-Driven Antibody Design
The Application of Artificial Intelligence in Antibody Design With the introduction of AI technology, the field of antibody discovery has undergone a revolutionary transformation. AI, through deep learning and machine learning algorithms, utilizes its ability to process and analyze biological big data to quickly identify and predict potential antibody sequences, greatly improving the discovery efficiency and success rate.
Experimental Validation of AI-Designed Antibodies
The final validation of the efficacy and safety of antibodies designed with the assistance of Artificial Intelligence (AI) is a crucial step. Laboratory validation involves not only confirming whether the antibody can specifically recognize the target antigen but also assessing its bioactivity and performance within a biological organism. This process faces a series of challenges and technical difficulties.
The preliminary validation of antibody design typically begins with testing its affinity and specificity. This involves a series of biochemical experiments, such as Enzyme-Linked Immunosorbent Assay (ELISA), Surface Plasmon Resonance (SPR), and Flow Cytometry, to ensure that the antibody can specifically bind to its target antigen.
After confirming the antibody's affinity and specificity, the next step is to assess its functionality, including testing the antibody’s effect on target cells in in vitro models or evaluating its therapeutic effects in animal models. Simultaneously, safety assessment is indispensable, especially before clinical application. This includes evaluating whether the antibody might trigger adverse immune reactions, toxicity, or other potential side effects.
Finally, AI-designed antibodies must undergo a series of comprehensive evaluations, including production costs, stability, preparation difficulty, and batch-to-batch consistency. These factors collectively determine whether the antibody can transition from the laboratory to clinical application.
Gene Universal High-Throughput Antibody Expression Platform
In the field of antibody development, Gene Universal stands out with its high-throughput expression platform, "from gene sequence to antibody," providing a standardized, high-throughput antibody production process that offers a highly cost-effective solution for large-scale screening of recombinant antibodies.
By simply providing the DNA sequence, the full process from DNA sequence to purified antibody can be completed and delivered in as little as one week, significantly shortening the antibody development cycle. Paired with the gold-standard Biacore affinity testing, this platform contributes to advancing AI-driven next-generation antibody discovery!
Service Advantages
Recombinant Antibody Expression Service
Fast Delivery in One Week - No Success, No Charge
|
Expression Type
|
Expression System |
QC |
Turnaround |
Deliverable |
|
|---|---|---|---|---|---|
|
Standard |
Fast |
||||
|
Recombinant Antibodies (IgGs, Fab, scFv, VHH, bispecific antibodies, etc.) |
GUeasy 293/CHO |
►SDS-PAGE > 95% |
2 Weeks |
1 Week |
►Expression plasmid contain GOI |
Note: Smaller or larger expression volumes can be selected based on requirements
Case Study
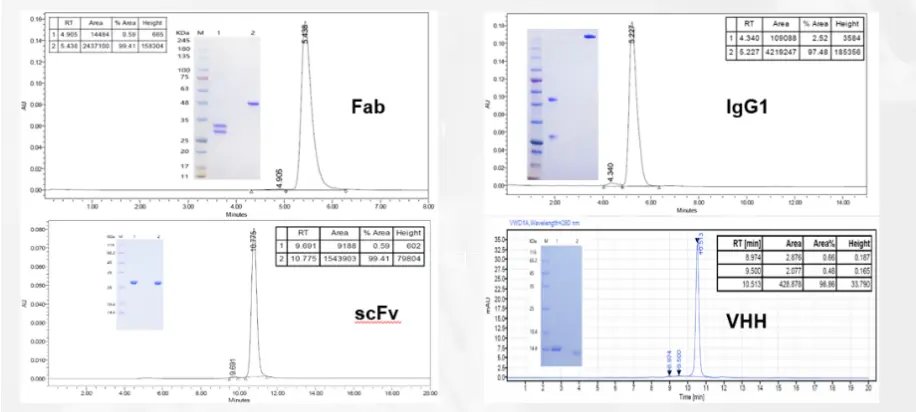
Antibody Affinity Measurement Service
Biacore T200 Facilitates Preliminary Validation of Antibody Design
Preliminary validation of antibody design typically begins with testing its affinity and specificity. Biacore can be used to analyze the binding and dissociation constants of different antibodies with antigens or proteins with small molecules. As a comprehensive platform for antibody bioactivity testing, its accurate and stable data quality has been widely applied and recognized by pharmaceutical companies and regulatory agencies.
Gene Universal offers a SPR-based biomolecular interaction analysis system (Biacore T200) service platform, which provides advantages such as label-free samples, high sensitivity, rapid detection, and real-time quantitative testing. This platform can perform antibody affinity measurement services and provide comprehensive interaction data between antibody samples and specific molecules.

